 Every now and then science comes up with a new approach to research that impacts on technology, but often these approaches are controversial and the headlines we see are far from the truth and can damage the investment into the new techniques. One good example is the Genetic Modification of plants and the production of GM-foods, which has a really bad press in Europe despite many obvious benefits for the world economy and for disease control. The latest technology, which follows from the explosion in genetic engineering techniques during the 1990s, builds on concepts developed in bionanotechnology and is known as Synthetic Biology. But, what is Synthetic Biology? Will it work? And what are the dangers versus benefits of these developments? Gardner and Hawkins (2013) have written a recent review about this subject, which made me think a blog on the subject was overdue.
Every now and then science comes up with a new approach to research that impacts on technology, but often these approaches are controversial and the headlines we see are far from the truth and can damage the investment into the new techniques. One good example is the Genetic Modification of plants and the production of GM-foods, which has a really bad press in Europe despite many obvious benefits for the world economy and for disease control. The latest technology, which follows from the explosion in genetic engineering techniques during the 1990s, builds on concepts developed in bionanotechnology and is known as Synthetic Biology. But, what is Synthetic Biology? Will it work? And what are the dangers versus benefits of these developments? Gardner and Hawkins (2013) have written a recent review about this subject, which made me think a blog on the subject was overdue.
My background in this area is two-fold:
- I was a part of a European Road-Mapping exercise, TESSY, that produced a description of what Synthetic Biology is and how it should be implemented/funded in Europe.
- I was also Project Coordinator for a European research project – BioNano Switch, funded by a scheme to support developments in Synthetic Biology, that aimed to produce a biosensor using an approach embedded in the concepts of Synthetic Biology.
So, what is Synthetic Biology? I think the definition of this area of research needs to be clearly presented, something that was an important part of the TESSY project, as the term has become associated simply with the production of an artificial cell. However, that is only one small aspect of the technology and the definition TESSY suggested is much broader:
Synthetic Biology aims to engineer and study biological systems that do not exist as such in nature, and use this approach for:
-
achieving better understanding of life processes,
-
generating and assembling functional modular components,
-
developing novel applications or processes.
 This is quite a wide definition and is best illustrated with a simple comparison – in electronic engineering there exists a blueprint (circuit diagram) that shows how components (resistors, capacitors etc.) can be fitted together in a guaranteed order to produce a guaranteed result (a device such as an amplifier). The Synthetic Biology concept would be to have a collection of such components (DNA parts that include promoters, terminators, genes and control elements; cellular systems including artificial cells and genetically engineered bacteria capable of controlled gene expression; interfaces that can connect biological systems to the human interface for useful output). This would mimic the electronic situation and provide a rapid mechanism for assembly of biological parts into useful devices in a reliable and predictable manner. There are many examples of such concepts, but the best known is the Biobricks Foundadtion. However, at the TESSY meeting I was keen to make it clear that there are fundamental problems with this concept, so what are the problems?
This is quite a wide definition and is best illustrated with a simple comparison – in electronic engineering there exists a blueprint (circuit diagram) that shows how components (resistors, capacitors etc.) can be fitted together in a guaranteed order to produce a guaranteed result (a device such as an amplifier). The Synthetic Biology concept would be to have a collection of such components (DNA parts that include promoters, terminators, genes and control elements; cellular systems including artificial cells and genetically engineered bacteria capable of controlled gene expression; interfaces that can connect biological systems to the human interface for useful output). This would mimic the electronic situation and provide a rapid mechanism for assembly of biological parts into useful devices in a reliable and predictable manner. There are many examples of such concepts, but the best known is the Biobricks Foundadtion. However, at the TESSY meeting I was keen to make it clear that there are fundamental problems with this concept, so what are the problems?
At its most simple concepts a Biobricks database would consists of a number of different types of DNA (promoters, are short DNA sequences that switch a gene on; terminators, are short DNA sequences that switch a gene off; control elements, are DNA sequences that control the promoter switching on or off a gene as required; genes, would be DNA sequences that produce 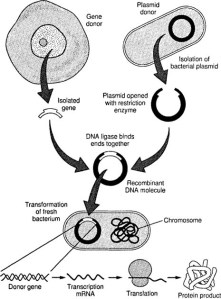 biotechnologically useful products; and cells, are the final package that enables the DNA to do its work and produce the required product), which sounds logical and quite simple. However, biological systems are not as reliable as electronic systems and combinations of promoters and genes do not always work. One of the major problems with protein production, using such artificial recombinant systems, is protein aggregation resulting in insoluble proteins that are non-functional. In addition, there are many examples (usually unpublished) of combinations of Biobricks that do not work as expected, or if used in a different order also result in protein aggregation, none of which ever happens with electronic components. The reasons are far from clear, but are closely related to the complexity of proteins and the need for them to operate in an aqueous environment. My thoughts about how to deal with this situation is to have a large amount of metadata associated with any database of Biobricks, which includes information about failures or problems of protein production from specific combinations. However, I am not aware of any such approach!
biotechnologically useful products; and cells, are the final package that enables the DNA to do its work and produce the required product), which sounds logical and quite simple. However, biological systems are not as reliable as electronic systems and combinations of promoters and genes do not always work. One of the major problems with protein production, using such artificial recombinant systems, is protein aggregation resulting in insoluble proteins that are non-functional. In addition, there are many examples (usually unpublished) of combinations of Biobricks that do not work as expected, or if used in a different order also result in protein aggregation, none of which ever happens with electronic components. The reasons are far from clear, but are closely related to the complexity of proteins and the need for them to operate in an aqueous environment. My thoughts about how to deal with this situation is to have a large amount of metadata associated with any database of Biobricks, which includes information about failures or problems of protein production from specific combinations. However, I am not aware of any such approach!
 There are other aspects of Synthetic Biology that do not depend on Biobricks and one example is the artificial cell. The ideal for such a system is a self-assembling package, capable of entrapping DNA, capable of replication and survival and able to produce useful biomaterials and significant steps have been made toward such a system. However, one area of concern as such systems are developed, is containment – can we really be sure these artificial microbes will remain in a contained environment and not escape to interact with and possible change the natural bacterial population. However, the power and capability of such a system should not be underestimated and the likely use in future medicine could be immense – simple examples would be as delivery systems for biomaterial that can activate cellular changes by targeting to the required cell and then switching on protein production (e.g. hormones). This type of targeted medicine would be a major breakthrough during the later part of this century.
There are other aspects of Synthetic Biology that do not depend on Biobricks and one example is the artificial cell. The ideal for such a system is a self-assembling package, capable of entrapping DNA, capable of replication and survival and able to produce useful biomaterials and significant steps have been made toward such a system. However, one area of concern as such systems are developed, is containment – can we really be sure these artificial microbes will remain in a contained environment and not escape to interact with and possible change the natural bacterial population. However, the power and capability of such a system should not be underestimated and the likely use in future medicine could be immense – simple examples would be as delivery systems for biomaterial that can activate cellular changes by targeting to the required cell and then switching on protein production (e.g. hormones). This type of targeted medicine would be a major breakthrough during the later part of this century.
 Another type of Synthetic Biology involves the artificial assembly (possible self assembly) of biomaterials onto an artificial surface in an way that is unlikely to occur naturally, but provides a useful device – I see this as more like what a Biobricks project should be like – such a system is usually modular in nature and the bio-material would normally be produced using recombinant techniques. The research project I mentioned earlier involved such a device and the outcome was a single molecule biosensor for detecting drug-target interactions at the limits of sensitivity. The major issues we had with developing this device was the precise and accurate attachment of biomaterial, to a surface in such a way that they function normally. However, overall the project was successful and shows that a Synthetic Biology approach has merits.
Another type of Synthetic Biology involves the artificial assembly (possible self assembly) of biomaterials onto an artificial surface in an way that is unlikely to occur naturally, but provides a useful device – I see this as more like what a Biobricks project should be like – such a system is usually modular in nature and the bio-material would normally be produced using recombinant techniques. The research project I mentioned earlier involved such a device and the outcome was a single molecule biosensor for detecting drug-target interactions at the limits of sensitivity. The major issues we had with developing this device was the precise and accurate attachment of biomaterial, to a surface in such a way that they function normally. However, overall the project was successful and shows that a Synthetic Biology approach has merits.
What are the benefits that Synthetic Biology can provide society? Well, one advantage is a more systematic approach to biotechnology, which to date has tended to move forward at the whim of researchers in Academia or industry. Assuming the problems, associated with protein production, mentioned above can be better understood then there could be a major boost in use of proteins for biotechnology. In addition, Synthetic Biology techniques offer a unique opportunity for miniaturisation and mass production of biosensors that could massively improve medical diagnosis. Finally, artificial cells have many future applications in medicine, if they can be produced in a reliable way and made to work as expected:
- They could provide insulin for diabetics.
- Be made to generate stem cell, which could be used in diseases such as Alzheimer’s and Huntingdon’s.
- They could deliver specific proteins, drugs and hormones to target locations.
- They could treat diseases that result from faulty enzyme production (e.g. Phenylketonuria).
- They could even be used to remove cholesterol from the blood stream.
However, there are always drawbacks and risks associated with any new scientific advance:
- Containment of any artificial organism is the most obvious, but this enhanced by the possibility of using the organism to produce toxins that would allow its use as a biological weapon.
- The ability to follow a simple “circuit diagram” for protein production, combined with a readily available database of biological material, could enable a terrorist to design a lethal and unpredictable weapon much more complex and perhaps targeted than anything known to date.
- Inhibit research through a readily available collection of materials that prevent patent protection of inventions. This could be complicated by the infringement of patents by foreign powers in a way that blocks conventional research investment.
- Problems associated with the introduction of novel nano-sized materials into the human body, including artificial cells, which may be toxic in the long term.
My own feeling is that we must provide rigorous containment and controls (many of which already exist), but allow Synthetic Biology to develop, Perhaps there should be a review of the situation by the end of this decade, but I hope that the risks do not materialise and that society can benefit from this work.
 I was reading an article in Scientific American today that got me thinking about the complexities of biology – the article described the production of a virtual bacteria using computing to model all of the known functions of a simple single cell. The article was a very compelling read and presented a rational argument of how this could be achieved, based on modelling of a single bacterium that would eventually divide. The benefits of a successful approach to achieving this situation are immense for both healthcare and drug development, but the difficulties are equally immense.
I was reading an article in Scientific American today that got me thinking about the complexities of biology – the article described the production of a virtual bacteria using computing to model all of the known functions of a simple single cell. The article was a very compelling read and presented a rational argument of how this could be achieved, based on modelling of a single bacterium that would eventually divide. The benefits of a successful approach to achieving this situation are immense for both healthcare and drug development, but the difficulties are equally immense. The number of copies of a plasmid are tightly controlled in a bacterial cell and this control is usually governed by an upper limit to an encoded RNA or protein, when the concentration of this product drops, such as AFTER division of the cell, replication will occur and the number of plasmids will increase until the correct plasmid number is attained (indicated by the concentration of the controlling gene product).
The number of copies of a plasmid are tightly controlled in a bacterial cell and this control is usually governed by an upper limit to an encoded RNA or protein, when the concentration of this product drops, such as AFTER division of the cell, replication will occur and the number of plasmids will increase until the correct plasmid number is attained (indicated by the concentration of the controlling gene product).

















You must be logged in to post a comment.