 When I first diagnosed with Type 2 Diabetes (TD2) I immediately started to research the scientific literature for any clear genetic explanation of this highly prevalent disease, but I was unable to find any clear link between a specific genetic loci and occurrence of the disease; although, there were several papers making such links they were far from proving any such link. However, a recent article in Scientific American (October 2015, pp56-59) has suggested one possible explanation of the growth of TD2 and a genetic cause that predates the evolution of Homo Sapiens! Reflecting on this article I can understand how I would not have come across this explanation as the research has always been linked to a different disorder – Gout, or the “disease of Kings”.
When I first diagnosed with Type 2 Diabetes (TD2) I immediately started to research the scientific literature for any clear genetic explanation of this highly prevalent disease, but I was unable to find any clear link between a specific genetic loci and occurrence of the disease; although, there were several papers making such links they were far from proving any such link. However, a recent article in Scientific American (October 2015, pp56-59) has suggested one possible explanation of the growth of TD2 and a genetic cause that predates the evolution of Homo Sapiens! Reflecting on this article I can understand how I would not have come across this explanation as the research has always been linked to a different disorder – Gout, or the “disease of Kings”.
Gout is caused by a build up of uric acid in the bloodstream, which can then crystallise in capillary vessels leading to immense pain. Uric acid is swiftly removed from most animals through breakdown by an enzyme called Uricase, but humans and many primates lack a functional form of the gene responsible for production of this enzyme. Apparently, the loss of function of this gene occurred some 15+ million years ago when a series of nonsense mutations inactivated the gene (Oda et al. Mol Biol Evol 2002;19:640–53). The article proposes that the selective pressure for the loss of Uricase activity begins when apes moved from Africa to Europe, which at first provided a plentiful environment with a sub-tropical climate providing bountiful supplies of fruit for their diet (particularly figs).
However, this period saw the beginning of climate cooling and this drier cooler weather changed the European vegetation from a rich broadleaf forested area toward a savanna-like environment, with much less fruit available and much of this fruit (especially figs)now becoming seasonal and quite scarce during winter. As cooling continued these European apes began to starve, therefore, the loss of the Uricase gene must have provided a selective advantage (Hayashi et al. Cell Biochem Biophys 2000;32:123–9). The normal mammalian reaction to periods of starvation is to produce fat (e.g. for an energy supply during hibernation, or to provide sufficient energy to survive winters). However, during prolonged periods of starvation foraging for food must continue, especially for primates that do not hibernate, and for this to be successful glucose is required by the brain. This is achieved by an “insulin-resistance” effect. The clue to this selective advantage lies with the fruit-rich diet that the apes in both Europe and Africa were consuming – digestion of fructose leads to production of uric acid and researchers have found that uric acid can trigger this switch to “insulin-resistance”. The researchers proposal is that the loss of the Uricase gene led to a gradual development of the ability to switch to converting fructose to fat providing a better chance to survive food shortages during winter. They also propose that these European apes may have brought this major selective advantage back to Africa as they migrated back to avoid cooling winters, they must have out-competed African apes and thus left the mutated Uricase gene that has been acquired by humans.
The researchers proposal is that the loss of the Uricase gene led to a gradual development of the ability to switch to converting fructose to fat providing a better chance to survive food shortages during winter. They also propose that these European apes may have brought this major selective advantage back to Africa as they migrated back to avoid cooling winters, they must have out-competed African apes and thus left the mutated Uricase gene that has been acquired by humans.
If this explanation of these genetic events is correct, we have a genetic explanation of TD2 – sometimes known as insulin-resistance – and what we have now is that processed foods, which often contain corn syrup, or table sugar, that are extremely rich in fructose, are being turned into fat because of the elevated uric acid levels in our bloodstream. It would be exciting to think that new drugs could be developed against uric acid production, which might help reduce obesity and TD2. Genetic Engineering may even hold the possibility of restoring Uricase production in the distant future. In the meantime, as I have said before we must aim to increase regular exercise, reduce sugar intake and aim to make fresh fruit our only supply of fructose. The antioxidants available in fresh fruit help to reduce many side effects of excess uric acid and reduce multiple diseases.
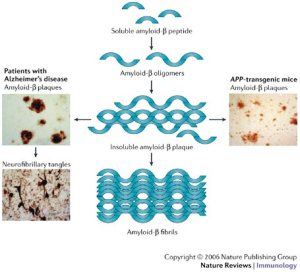
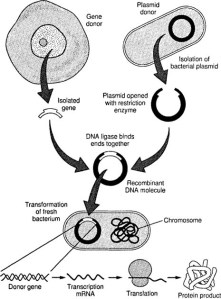
However, from a personal viewpoint I am left with something of a mystery as this genetic explanation does not explain familial occurrences of TD2, something I have personal experience of! The best link between TD2 occurrences in families and an observed disorder is that TD2 is tightly linked to β-cell dysfunction in the pancreas (O’Rahilly, S.P. et al. The Lancet , Volume 328 , Issue 8503 , 360 – 364), which is associated with insulin resistance (Kahn, 2003. Diabetologia 46, 3-19), but the nature of this genetic link is complex and confused and involves amyloidosis of insulin. A detailed description of this will follow.






















 The above title is
The above title is 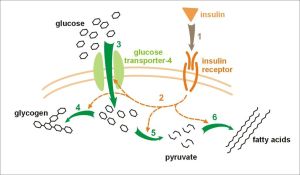







![nanopore_x616[1]](https://firmank.files.wordpress.com/2013/06/nanopore_x6161.jpg?w=293&h=300)
You must be logged in to post a comment.